Why doesn't FemINDICAtor® use the published Mandolino Primer sets?
Next-generation sequencing revealed polymorphisms within the Mandolino-published MADC2 regions, making them problematic targets for qPCR.
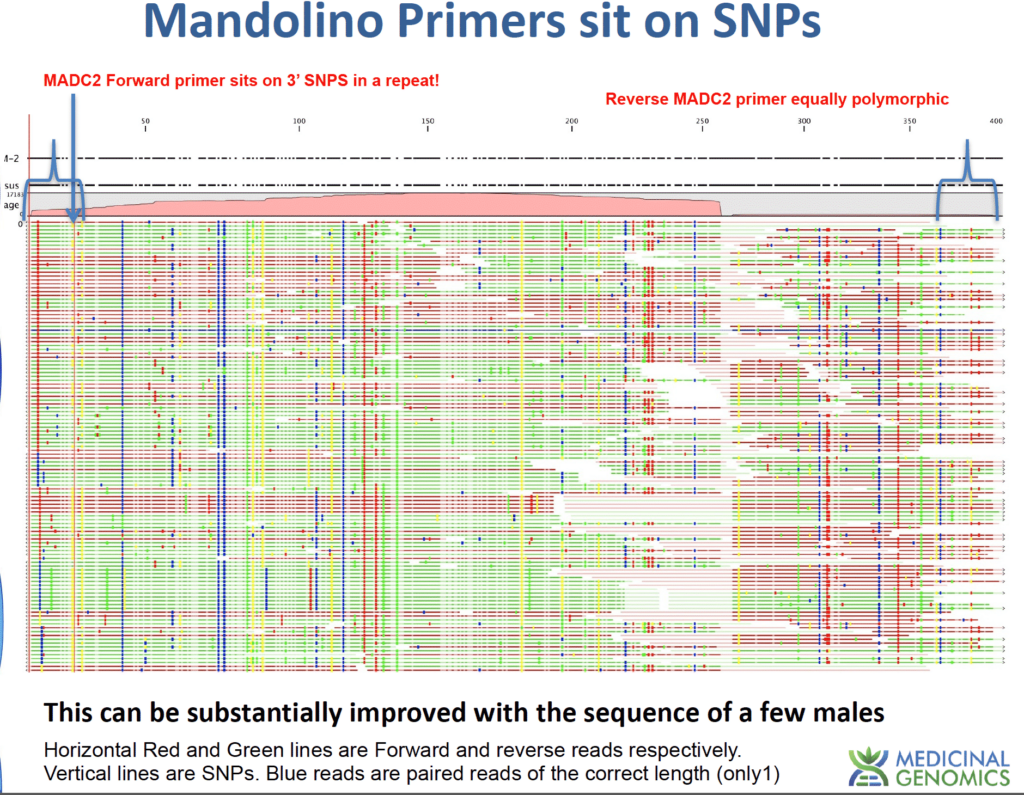
Further sequencing of the Mandolino-published MADC2 regions shows these regions are problematic targets for qPCR. Specifically, the Mandolino primer sets target a repeat in the genome that is not unique to males. A single-base variant in this region provides one extra band for most males. Most males is the key issues here. When failure is not an option repeats with 500 copies in the female genome cannot be part of a qPCR product offering to identify males. Too little is know about these mobile elements to bet on them for large crops at risk of pollination. Ask your test provider what they target and how they validated their male test before they blame you for “hermi”ing your plants.
Below is a depiction of the MADC2 region after mapping reads to the loci. This was a 30X male genome that ended up with 17,000X coverage over the MADC2 loci and many polymorphisms (vertical lines) with very few paired reads agreeing on the full-length nature of the repeat. This explains the complicated banding patterns described in the Mandolino et al paper. The blue brackets below represent the primer locations. The polymorphisms under the primers are a concern for qPCR assays.