Why is Real-time PCR is superior to end-point PCR?
Real-time or quantitative PCR (qPCR) provides more consistent, reproducible results and is more sensitive than end-point PCR
To understand the difference between end-point PCR and real-time PCR, one first must understand what happens during a PCR reaction.
PCR amplifies a segment of target DNA to produce millions of copies – sometimes from just a single copy. This process is carried out over a series of heating cycles, and as a result, a basic PCR run can be broken up into three phases:
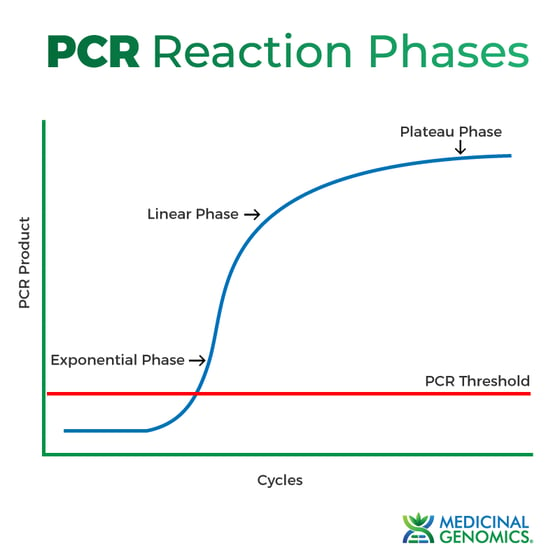
Figure 1: PCR phases.
Exponential
During this phase, the target DNA doubles with each cycle (assuming 100% reaction efficiency). Exponential amplification occurs because all of the reagents are fresh and available, and the kinetics of the reaction push the reaction to favor doubling.
Linear (high variability)
At this point, reagents are becoming scarce and PCR product is not doubling each cycle.
Plateau (End-point: gel detection for traditional methods)
When the line plateaus the reaction has stopped, and no more PCR products are being made. This is where end-point PCR takes its measurement. However, even reactions with the same starting DNA concentration will plateau at a different point, due to the different reaction kinetics for each sample.
Traditional PCR measures at the plateau, giving you variable results
Figure 2 clearly shows variations in the plateau phase of multiple PCR reactions with the same starting DNA concentration. In this example, 96 replicate reactions that were run on the same plate. Note that the fluorescence values at the endpoint show a lot of variability from sample to sample. This is due to variations in the reaction kinetics.
Real-time PCR measures at the exponential phase for more accurate quantitation
Real-time PCR focuses on the exponential phase because it provides the most precise and accurate data for quantitation. Within the exponential phase, the real-time PCR instrument determines at which cycle a reaction reaches a fluorescent intensity above background. This level is called the Cycle Quantification, Cq.
In Figure 2, you can see the reactions all show very tight agreement where the plots cross the Threshold, making that value much more consistent.
The Cq value is used in downstream quantitation or presence/absence detection. By comparing the Cq values of samples of unknown concentration with a series of standards, the amount of template DNA in an unknown reaction can be accurately determined.
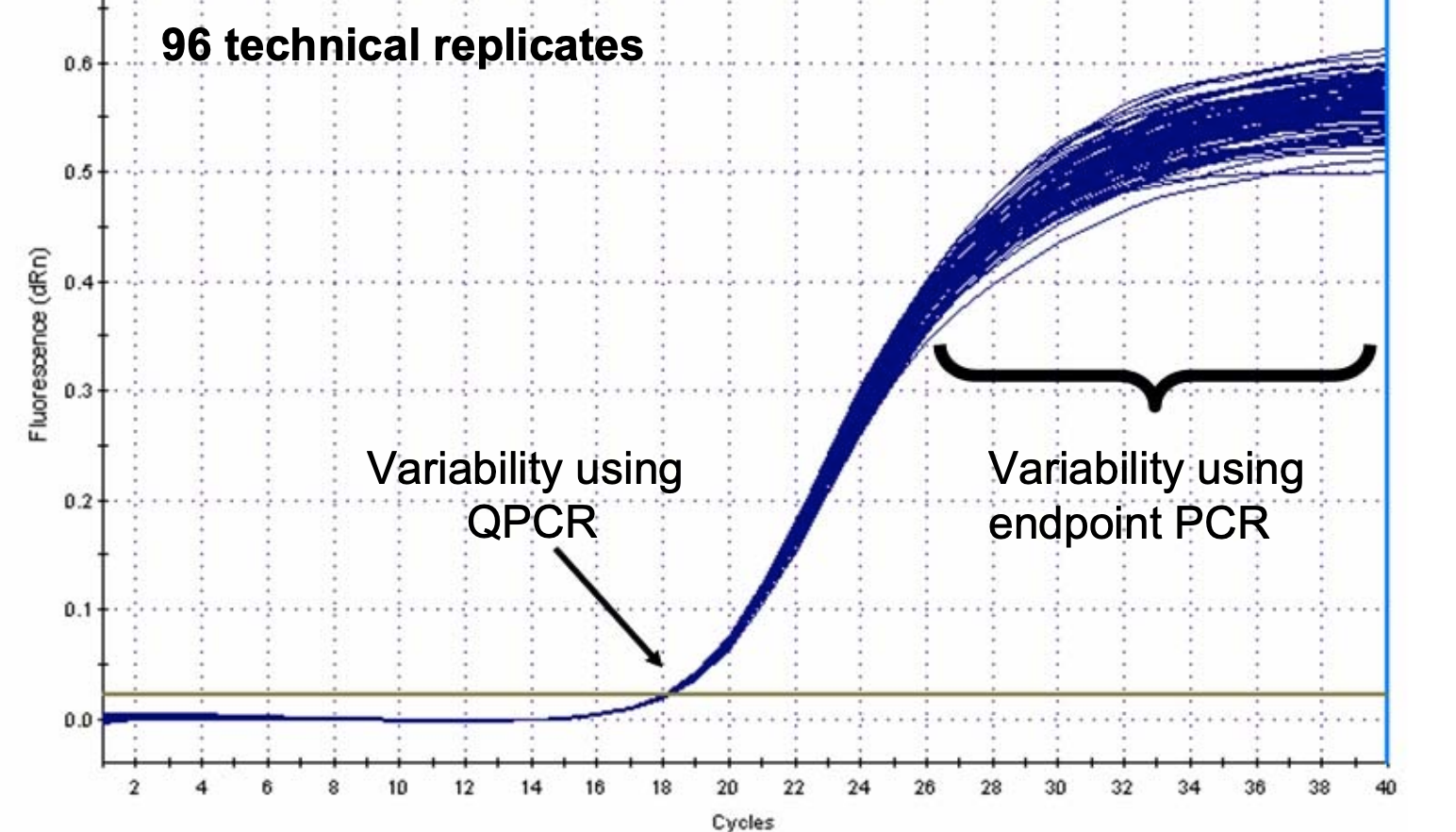
Figure 2: Identical samples produce different quantities of reaction products by the plateau phase of PCR while Cq values remain consistent.
qPCR probes add an additional level of sensitivity
All PCR reactions use primers, which are short DNA sequences that determine what part of the target DNA will be amplified. Primers are designed to bind adjacent to the target sequence and are specific to the target DNA. If even a single DNA base is different, binding may not occur and the DNA sequence will not be amplified.
qPCR also uses primers, which are small DNA sequences that have a fluorescent dye attached. Those primers attach a portion of the target DNA sequence, and when the DNA is amplified the probe emits a signal that is detected by the qPCR instrument. Primers provide an additional level of sensitivity because if they do not attach, the instrument will not detect the PCR product. This is useful when designing species-specific assays. In some cases, two qPCR assays may share a primer set, but use different probes to differentiate the species. End-point PCR does not have this additional level of specificity available.
Additional References
Real-Time vs. Digital PCR vs. Traditional PCR - Thermo Fisher Scientific
